Automatic Network Reconstruction
The idea behind Automatic Network Reconstruction is to have an algorithm that automatically gives all possible explanations of a set of experimental results or observations in terms of a complete list of networks of causal interactions between the measured components that can explain the experimental data. The completeness of the list is proven mathematically.
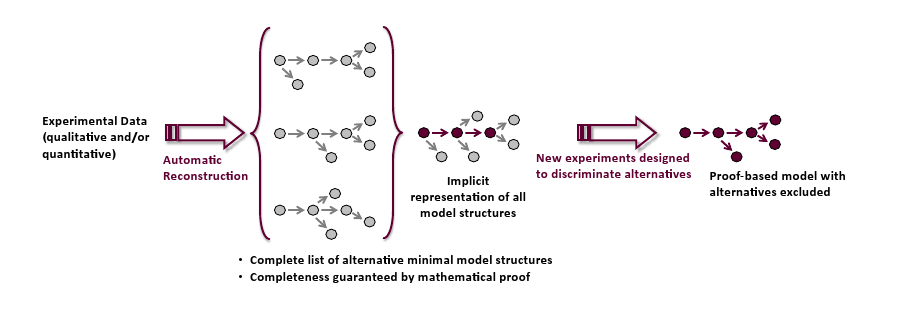
The alternative network structures found can be summarized in the form of an implicit representation telling which structural features all models have in common and which nodes of the network might be wired up alternatively as displayed. Considering the nodes with alternative connections one can design new experiments that specifically discriminate between the alternatives to finally obtain a model structure based on mathematical proof.
In cooperation with Annegret Wagler and Robert Weismantel we have developed the proof-based method and demonstrated its application to molecular biology. More recently, we have developed an algorithm for the reconstruction of extended Petri nets, described the underlying mathematics and its application to signal transduction and to gene regulatory networks. In cooperation with Torsten Schaub we have shown that Answer Set Programming is an appropriate method for solving the reconstruction problem in a computationally efficient way. The next step is to try the method with real experimental data.
Using Snoopy, a tool for Advanced Petri Nets which is developed by Monika Heiner and her group, the reconstructed networks can be displayed as Petri nets and simulated as continuous (ODE-based) or stochastic models. Snoopy runs under Linux, Mac OS X and Windows.