Petri Nets
The formal language of Petri nets offers a unifying and powerful framework for modeling in systems and synthetic biology. Inherently concurrent, asynchronous and nondeterministic processes which are typical for biochemical reaction networks and molecular mechanisms in general are unambigously described with a strict syntax. The resulting graphical representation relies on only few symbols and is intuitively understandable. The graph of a Petri net is a well-defined mathematical structure with operational semantics. This means that a Petri net describing a (molecular) mechanism is equivalent to a protocol which is executable on an abstract machine or on a real computer. Simultaneously, a given Petri net graph can be automatically interpreted as qualitative, continuous, stochastic or hybrid model (what you see is what you get; WYSIWYG) and simulations directly run. Petri nets support hierarchical concepts as well as modular approaches. The colored extension of low-level Petri nets combines the strength of Petri nets with the expressive power of programming languages. This extension defines datatypes and algorithms to create models with complex structure. This feature is especially helpful for the generation of multiscale models e.g. for modeling a population of identical or modified reactions networks distributed over individual cells. The Petri net formalism is supported by several powerful modeling, analysis and simulation tools, Snoopy, Charlie, Marcie developed by Monika Heiner and her group (free download at: Brandenburg University of Technology at Cottbus). Petri net tools like Snoopy are capable of importing and exporting SBML to be compatible with other modeling frameworks common in systems biology. Snoopy runs under Linux, Mac OS X and Windows.
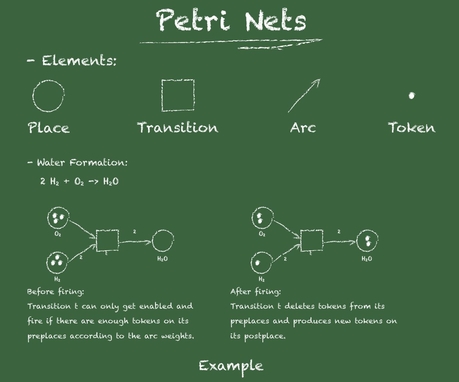
Fig. 1 Petri net formalism.
References on how to use Petri nets and Snoopy
We have written a couple of tutorial book chapters on the use of Petri nets and Snoopy for modeling and simulation of biochemical or gene regulatory networks that may help to get started.
Basic
Marwan W., C. Rohr and M. Heiner. 2012. Petri nets in Snoopy: A unifying framework for the graphical display, computational modelling, and simulation of bacterial regulatory networks in Methods in Molecular Biology, Springer 804/2012:409-437.
Rohr C., W. Marwan and M. Heiner. 2010. Snoopy - a unifying Petri net framework to investigate biomolecular networks. Bioinformatics, 26,7:974-975.
Advanced
Blätke M.A., M. Heiner and W. Marwan. 2015. BioModel Engineering with Petri Nets. Algebraic and Discrete Mathematical Methods for Modern Biology (ed. R Robeva), pp. 141-192. Academic Press, Burlington.
Blätke M.A., C. Rohr, M. Heiner and W. Marwan. 2014. A Petri-Net-Based Framework for Biomodel Engineering. In Large-Scale Networks in Engineering and Life Sciences (pp. 317-366). Springer International Publishing.
Blätke, M.A. 2011. Tutorial Petri Nets in Systems Biology. Technical Report, Otto-von-Guericke University, Magdeburg